Make an initial guess of C4 hyperbola parameter values for one curve
initial_guess_c4_aci_hyperbola.RdCreates a function that makes an initial guess of C4 hyperbola model
parameter values for one curve. This function is used internally by
fit_c4_aci_hyperbola.
Values estimated by this guessing function should be considered inaccurate, and should always be improved upon by an optimizer.
Details
Here we estimate values of c4_curvature, c4_slope, rL,
and Vmax from a measured C4 CO2 response curve. For more information
about these parameters, see the documentation for
calculate_c4_assimilation_hyperbola.
Here we take a very simple approach to forming the initial guess. We always
choose c4_curvature = 0.5, c4_slope = 1.0, and rL = 0.0.
For Vmax, we use Vmax = max{A} - rL_guess, where max{A}
is the largest observed net CO2 assimilation rate and rL_guess is the
guess for rL.
Value
A function with one input argument rc_exdf, which should be an
exdf object representing one C4 CO2 response curve. The return value of
this function will be a numeric vector with four elements, representing the
values of c4_curvature, c4_slope, rL, and Vmax
(in that order).
Examples
# Read an example Licor file included in the PhotoGEA package
licor_file <- read_gasex_file(
PhotoGEA_example_file_path('c4_aci_1.xlsx')
)
# Define a new column that uniquely identifies each curve
licor_file[, 'species_plot'] <-
paste(licor_file[, 'species'], '-', licor_file[, 'plot'] )
# Organize the data
licor_file <- organize_response_curve_data(
licor_file,
'species_plot',
c(9, 10, 16),
'CO2_r_sp'
)
# Create the guessing function
guessing_func <- initial_guess_c4_aci_hyperbola()
# Apply it and see the initial guesses for each curve
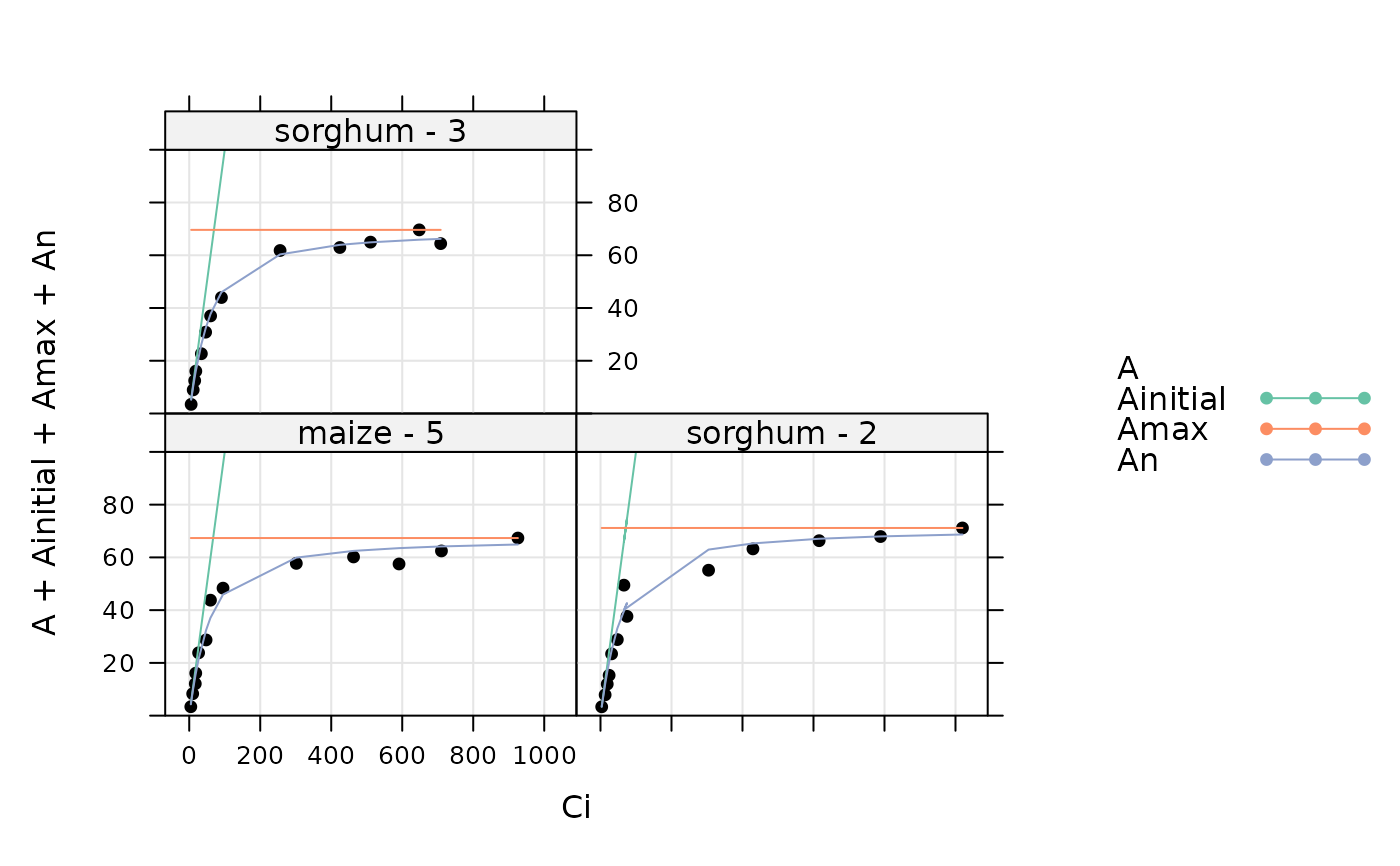
print(by(licor_file, licor_file[, 'species_plot'], guessing_func))
#> $`maize - 5`
#> [1] 0.50000 1.00000 0.00000 67.33821
#>
#> $`sorghum - 2`
#> [1] 0.50000 1.00000 0.00000 71.16098
#>
#> $`sorghum - 3`
#> [1] 0.50000 1.00000 0.00000 69.62463
#>
# A simple way to visualize the guesses is to "fit" the curves using the null
# optimizer, which simply returns the initial guess
aci_results <- consolidate(by(
licor_file,
licor_file[, 'species_plot'],
fit_c4_aci_hyperbola,
optim_fun = optimizer_null()
))
plot_c4_aci_hyperbola_fit(aci_results, 'species_plot', ylim = c(-10, 100))