Analyzing Ball-Berry Data
Source:vignettes/analyzing_ball_berry_data.Rmd
analyzing_ball_berry_data.RmdOverview
In this vignette, we will give an example showing how to analyze
Ball-Berry data using the PhotoGEA package. The commands in
this vignette can be used to initialize your own script, as described in
Customizing Your Script.
Background
The Model
The Ball-Berry model is a simple way to describe the response of a leaf’s stomata to its assimilation rate and local environmental conditions. Specifically, it predicts stomatal conductance to water vapor using the following equation:
where is the stomatal conductance, is the net assimilation rate, is the relative humidity at the leaf surface, and is the CO2 concentration at the leaf surface. The term is commonly referred to as the Ball-Berry index, and the model can be described as predicting a linear relationship between stomatal conductance and the Ball-Berry index. The intercept () and slope () of the linear relationship are referred to as the Ball-Berry parameters, and they describe the leaf’s stomatal response.
Although this model is certainly an oversimplification, it does encode some important stomatal responses. For example, when humidity is low, the stomata of a real plant tend to close, reducing stomatal conductance. Likewise, if the CO2 concentration around the leaf is depleted, the stomata of a real plant tend to open, increasing somatal conductance. For more information about this model and some possible alternatives, see the following papers:
The Data
Ball-Berry parameters are typically determined by measuring a Ball-Berry curve, where one or more of the environmental factors that influence the Ball-Berry index is systematically varied across a range of values. For each value, care is taken that and have reached their steady-state values, and then , , and other key pieces of information are recorded. Finally, a linear fit of the experimentally observed as a function of the Ball-Berry index is performed to extract values for the Ball-Berry intercept and slope.
Such photosynthetic gas exchange measurements are commonly performed
using a Licor portable photosynthesis system such as the Li-6400 or
Li-6800. These machines record values of
,
,
and many other important quantities. They produce two types of output
files: plain-text and Microsoft Excel. It is often more convenient to
work with the Excel files since the entries can be easily modified (for
example, to remove an extraneous row or add a new column). On the other
hand, it can be more difficult to access the file contents using other
pieces of software such as R. However, the PhotoGEA package
reduces this barrier by including tools for reading Licor Excel files in
R, which will be demonstrated in the following section.
Loading Packages
As always, the first step is to load the packages we will be using.
In addition to PhotoGEA, we will also use the
lattice package for generating plots.
If the lattice package is not installed on your R setup,
you can install it by typing
install.packages('lattice').
Loading Licor Data
The PhotoGEA package includes two files representing
Ball-Berry data measured using two Li-6800 instruments. The data is
stored in Microsoft Excel files, and includes curves measured from two
different crop species (tobacco and soybean) and several different plots
of each. Each curve is a seven-point light response curve; in other
words, the incident light intensity was varied to change
and hence the Ball-Berry index. Although these two files are based on
real data, noise was added to it since it is unpublished, so these files
should only be used as examples.
The files will be stored on your computer somewhere in your R package
installation directory, and full paths to these files can be obtained
with PhotoGEA_example_file_path:
# Define a vector of paths to the files we wish to load; in this case, we are
# loading example files included with the PhotoGEA package
file_paths <- c(
PhotoGEA_example_file_path('ball_berry_1.xlsx'),
PhotoGEA_example_file_path('ball_berry_2.xlsx')
)(Note: When loading your own files for analysis, it
is not advisable to use PhotoGEA_example_file_path as we
have done here. Instead, file paths can be directly written, or files
can be chosen using an interactive window. See Input Files below for more information.)
To actually read the data in the files and store them in R objects,
we will use the read_gasex_file function from
PhotoGEA. Since there are multiple files to read, we will
call this function once for each file using lapply:
# Load each file, storing the result in a list
licor_exdf_list <- lapply(file_paths, function(fpath) {
read_gasex_file(fpath, 'time')
})The result from this command is an R list of “extended data frames”
(abbreviated as exdf objects). The exdf class
is a special data structure defined by the PhotoGEA
package. In many ways, an exdf object is equivalent to a
data frame, with the major difference being that an exdf
object includes the units of each column. For more information, type
?exdf in the R terminal to access the built-in help menu
entry, or check out the Working With Extended Data
Frames vignette.
Generally, it is more convenient to work with a single
exdf object rather than a list of them, so our next step
will be to combine the objects in the list. This action can be
accomplished using the rbind function, which combines
table-like objects by their rows; in other words, it stacks two or more
tables vertically. This action only makes sense if the tables have the
same columns, so before we combine the exdf objects, we
should make sure this is the case.
The PhotoGEA package includes a function called
identify_common_columns that can be used to get the names
of all columns that are present in all of the Licor files. Then, we can
extract just those columns, and then combine the exdf
objects into a single one.
# Get the names of all columns that are present in all of the Licor files
columns_to_keep <- do.call(identify_common_columns, licor_exdf_list)
# Extract just these columns
licor_exdf_list <- lapply(licor_exdf_list, function(x) {
x[ , columns_to_keep, TRUE]
})
# Use `rbind` to combine all the data
licor_data <- do.call(rbind, licor_exdf_list)Now we have a single R object called licor_data that
includes all the data from several Licor Excel files. For more
information about consolidating information from multiple files, see the
Common Patterns section of the Working
With Extended Data Frames vignette.
Checking Licor Data
Before extracting Ball-Berry parameters, it is a good idea to do some basic checks of the data to ensure it is organized properly and that it was measured properly.
Basic Checks
First, we should make sure there is a column in the data whose value
uniquely identifies each curve. In this particular data set, several
“user constants” were defined while making the measurements that help to
identify each curve: instrument, species, and
plot. However, neither of these columns alone are
sufficient to uniquely identify each curve. We can solve this issue by
creating a new column that combines the values from each of these:
# Create a new identifier column formatted like `instrument - species - plot`
licor_data[ , 'curve_identifier'] <-
paste(licor_data[ , 'instrument'], '-', licor_data[ , 'species'], '-', licor_data[ , 'plot'])When measuring response curves (or any other type of gas exchange data), it is strongly recommended to record key pieces of metadata in the log files using “user constants,” as was done in this example. For more information, please see the Guide to Licor LI-6800 User Constants, which also includes advice for dealing with data sets that do not contain such metadata.
The next step is to make sure that this column correctly identifies
each response curve. To do this, we can use the
check_response_curve_data function from
PhotoGEA. Here we will supply the name of a column that
should uniquely identify each response curve
(curve_identifier), the expected number of points in each
curve (7), the name of a “driving” column that should
follow the same sequence in each curve (Qin), and tolerance
for the values of the driving column (1.0 mol / m^2 / s).
If the data passes the checks, this function will have no output and
will not produce any messages. (For more information, see the built-in
help menu entry by typing ?check_response_curve_data, or
visit the Frequently Asked
Questions.)
# Make sure the data meets basic requirements
check_response_curve_data(licor_data, 'curve_identifier', 7, 'Qin', 1.0)However, if check_response_curve_data detects an issue,
it will print a helpful message to the R terminal. For example, if we
had specified the wrong number of points, the wrong identifier column,
or an unrealistic tolerance, we would get error messages:
check_response_curve_data(licor_data, 'curve_identifier', 8)
#> curve_identifier npts
#> 1 ripe4 - soybean - 1a 7
#> 2 ripe4 - soybean - 1b 7
#> 3 ripe4 - soybean - 5 7
#> 4 ripe4 - tobacco - 2 7
#> 5 ripe5 - soybean - 1 7
#> 6 ripe5 - soybean - 5a 7
#> 7 ripe5 - soybean - 5c 7
#> 8 ripe5 - tobacco - 5 7
#> Error in check_response_curve_data(licor_data, "curve_identifier", 8): One or more curves does not have the expected number of points.
check_response_curve_data(licor_data, 'species', 7)
#> species npts
#> 1 soybean 42
#> 2 tobacco 14
#> Error in check_response_curve_data(licor_data, "species", 7): One or more curves does not have the expected number of points.
check_response_curve_data(licor_data, 'curve_identifier', 7, 'Qin', 0.1)
#> [1] "Point 1 from curve `ripe5 - soybean - 5a` has value `Qin = 2000.14`, but the average value for this point across all curves is `Qin = 2000`"
#> [2] "Point 1 from curve `ripe5 - soybean - 5c` has value `Qin = 2000.12`, but the average value for this point across all curves is `Qin = 2000`"
#> [3] "Point 2 from curve `ripe5 - soybean - 1` has value `Qin = 1100.11`, but the average value for this point across all curves is `Qin = 1099.965`"
#> [4] "Point 3 from curve `ripe5 - soybean - 1` has value `Qin = 800.223`, but the average value for this point across all curves is `Qin = 800.043375`"
#> [5] "Point 3 from curve `ripe5 - tobacco - 5` has value `Qin = 800.176`, but the average value for this point across all curves is `Qin = 800.043375`"
#> [6] "Point 4 from curve `ripe4 - soybean - 5` has value `Qin = 650.108`, but the average value for this point across all curves is `Qin = 650.005`"
#> [7] "Point 4 from curve `ripe5 - soybean - 1` has value `Qin = 650.115`, but the average value for this point across all curves is `Qin = 650.005`"
#> [8] "Point 5 from curve `ripe4 - soybean - 1a` has value `Qin = 500.135`, but the average value for this point across all curves is `Qin = 500.026875`"
#> [9] "Point 6 from curve `ripe5 - soybean - 1` has value `Qin = 350.145`, but the average value for this point across all curves is `Qin = 350.04`"
#> [10] "Point 7 from curve `ripe4 - tobacco - 2` has value `Qin = 200.236`, but the average value for this point across all curves is `Qin = 200.0565`"
#> Error in check_response_curve_data(licor_data, "curve_identifier", 7, : The curves do not all follow the same sequence of the driving variable.Qualitative Checks
Since the data has passed the basic checks, we can now take a look at
it to make sure it seems reasonable. Sometimes a Licor will override the
temperature or humidity controls while making measurements; in this
case, conditions inside the measurement chamber may not be stable, and
these curves may not accurately reflect the plant’s true steady-state
stomatal response. We can check for these types of issues by making
plots. In this situation, the lattice library makes it
simple to include each curve as its own separate subplot of a figure. In
the following sections, we will generate several different plots to
check each curve for quality.
Humidity Control
# Make a plot to check humidity control
xyplot(
RHcham + `Humidifier_%` + `Desiccant_%` ~ Qin | curve_identifier,
data = licor_data$main_data,
type = 'b',
pch = 16,
auto = TRUE,
grid = TRUE,
ylim = c(0, 100),
xlim = c(0, 2200),
xlab = paste0('Incident PPFD (', licor_data$units$Qin, ')')
)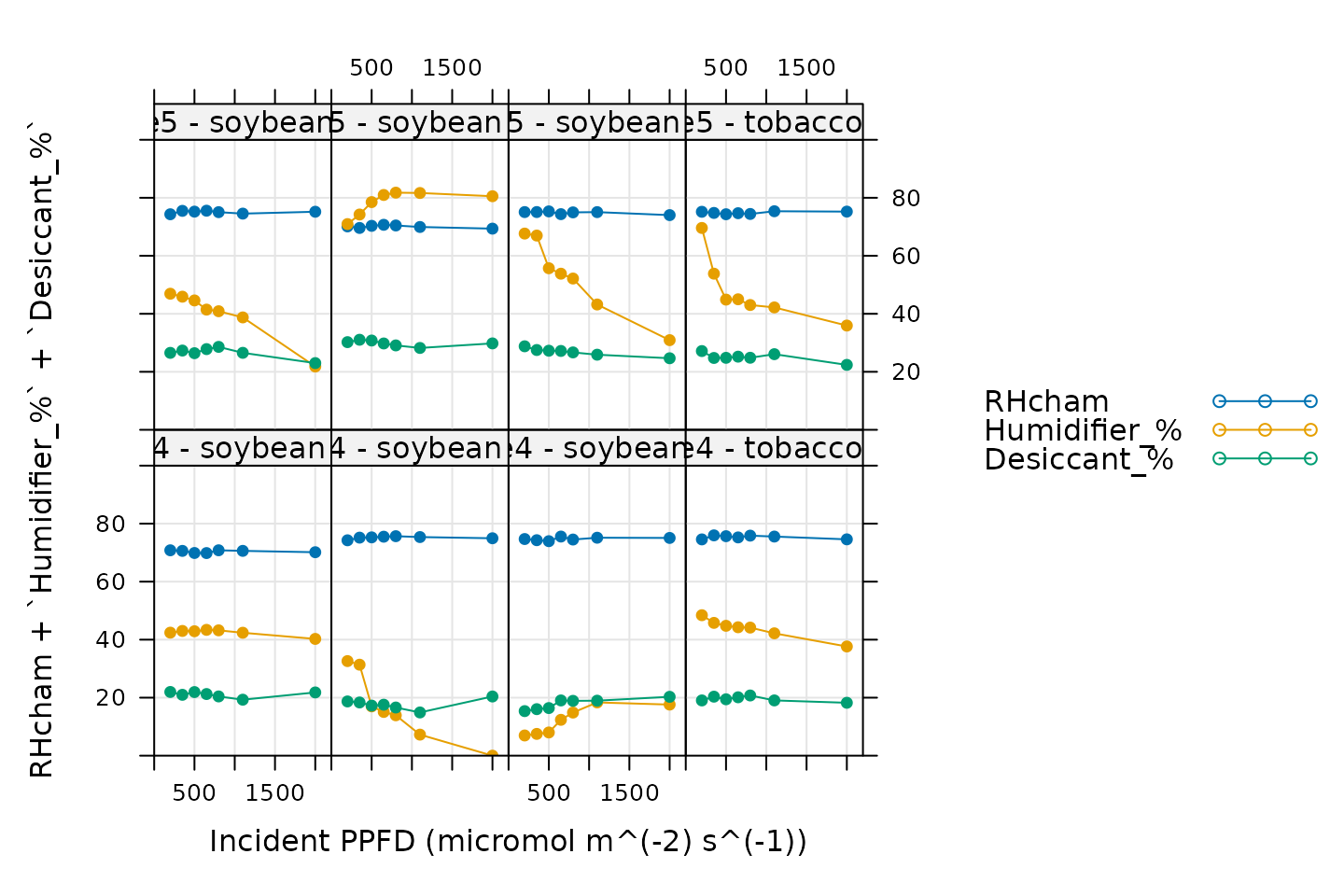
Here, Humidifier_% and Desiccant_%
represent the flow from the humidifier and desiccant columns, where a
value of 0 indicates that the valve to the column is fully closed and a
value of 100 indicates that the valve to the column is fully opened.
RHcham represents the relative humidity inside the chamber
as a percentage (in other words, as a value between 0 and 100).
Qin is the incident photosynthetically active flux density
(PPFD). When these curves were measured, a chamber humidity setpoint was
specified. So, when looking at this plot, we should check that the
relative humidity is fairly constant during each curve. Typically, this
should be accompanied by relatively smooth changes in the valve
percentages as they accomodate changes in ambient humidity and leaf
photosynthesis. In this plot, all the data looks good.
Temperature Control
# Make a plot to check temperature control
xyplot(
TleafCnd + Txchg ~ Qin | curve_identifier,
data = licor_data$main_data,
type = 'b',
pch = 16,
auto = TRUE,
grid = TRUE,
ylim = c(25, 40),
xlim = c(0, 2200),
xlab = paste0('Incident PPFD (', licor_data$units$Qin, ')'),
ylab = paste0('Temperature (', licor_data$units$TleafCnd, ')')
)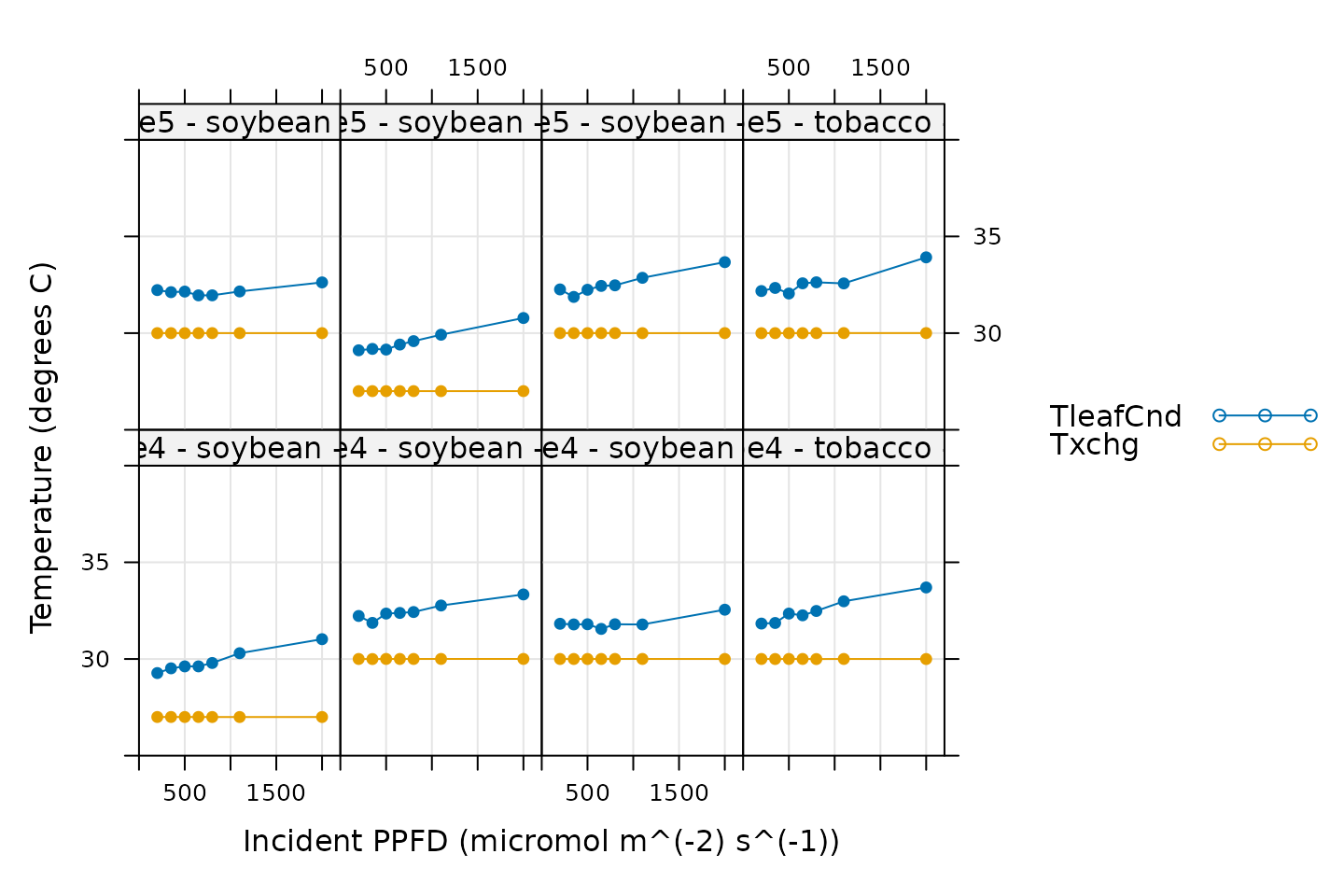
Here, TleafCnd is the leaf temperature measured using a
thermocouple, and Txchg is the temperature of the heat
exhanger that is used to control the air temperature in the measurement
instrument. When these curves were measured, an exchanger setpoint was
specified. So, when looking at this plot, we should check that
Txchg is constant during each curve and that the leaf
temperature does not vary in an erratic way. In this plot, all the data
looks good.
CO2 Control
# Make a plot to check CO2 control
xyplot(
CO2_s + CO2_r ~ Qin | curve_identifier,
data = licor_data$main_data,
type = 'b',
pch = 16,
auto = TRUE,
grid = TRUE,
ylim = c(360, 480),
xlim = c(0, 2200),
xlab = paste0('Incident PPFD (', licor_data$units$Qin, ')'),
ylab = paste0('CO2 concentration (', licor_data$units$CO2_r, ')')
)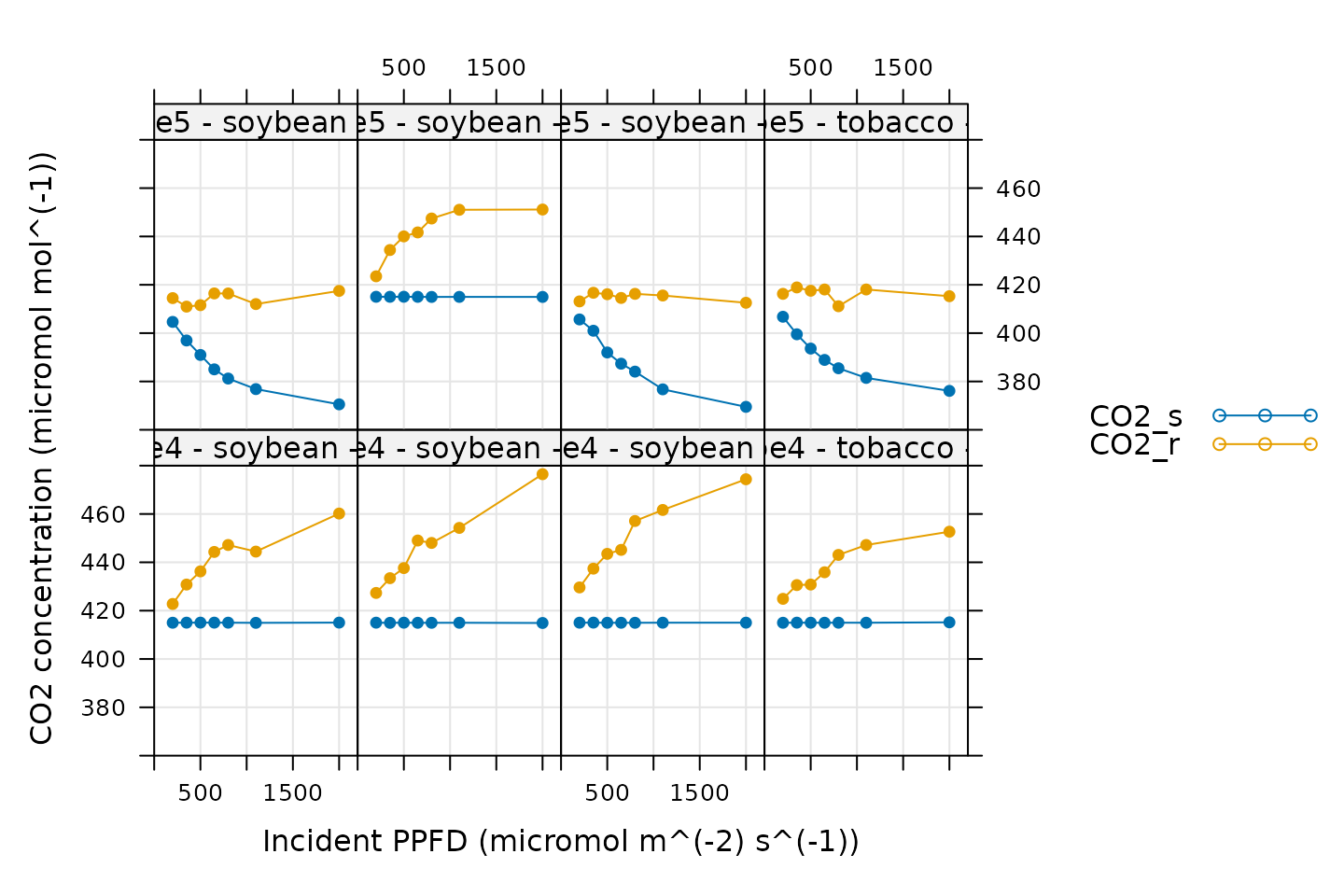
Here, CO2_s is the CO2 concentration in the
sample cell and CO2_r is the CO2 concentration
in the reference cell. When these curves were measured, a sample cell
CO2 concentration setpoint was supplied. So, when looking at
this plot, we should check that CO2_s is constant during
each curve. Here, it looks like the ripe5 instrument was
not controlling CO2 as expected during several of its curves;
CO2_s is not constant for ripe5 - soybean - 1,
ripe5 - soybean - 5c, and ripe5 - soybean - 5.
However, CO2_r is relatively constant during these curves,
and the changes in CO2_s are smooth, so it is reasonable to
expect that the measurements represent true steady-state values.
Considering this, all of these curves are acceptable based on the
CO2 plots.
Stability
xyplot(
`A:OK` + `gsw:OK` + Stable ~ Qin | curve_identifier,
data = licor_data$main_data,
type = 'b',
pch = 16,
auto = TRUE,
grid = TRUE,
xlim = c(0, 2200),
xlab = paste0('Incident PPFD (', licor_data$units$Qin, ')')
)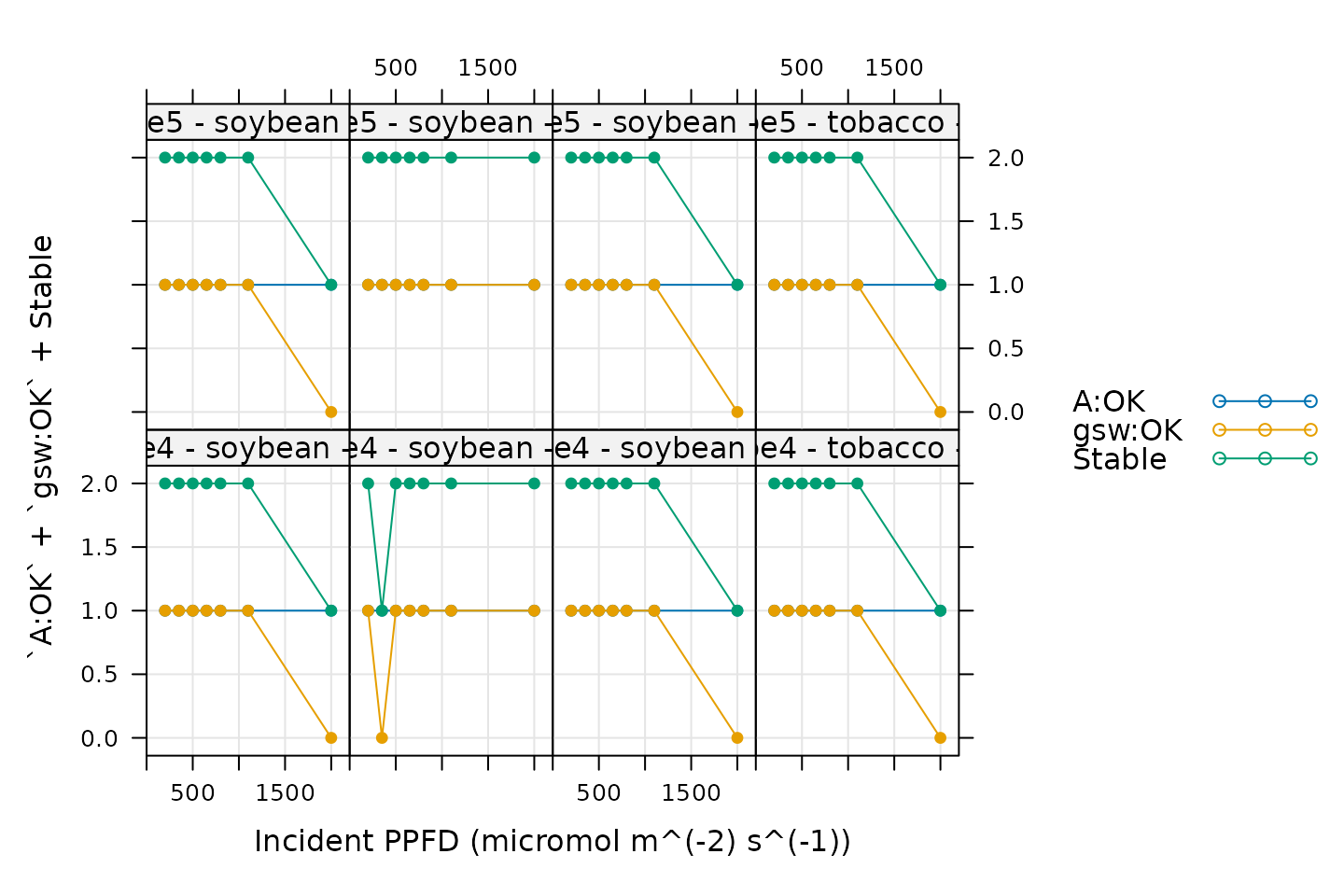
When measuring response curves with a Licor, it is possible to specify stability criteria for each point in addition to minimum and maximum wait times. In other words, once the set point for the driving variable is changed, the machine waits until the stability criteria are met; there is a minimum waiting period, and also a maximum to prevent the machine from waiting for too long. These stability criteria are especially important for Ball-Berry curves, since the stomata may take a long time to reach steady state.
When these curves were measured, stability criteria were supplied for
the net assimilation rate A and the stomatal conductance
gsw. The stability status for each was stored in the log
file because the appropriate logging option for stability was set. Now,
for each point, it is possible to check whether stability was achieved
or whether the point was logged because the maximum waiting period had
been met. If the maximum waiting period is reached and the plant has
still not stabilized, the data point may be unreliable, so it is very
important to check this information.
In the plot, A:OK indicates whether A was
stable (0 for no, 1 for yes), gsw:OK indicates whether
gsw was stable (0 for no, 1 for yes), and
Stable indicates the total number of stability conditions
that were met. So, we are looking for points where Stable
is 2. Otherwise, we can check the other traces to see whether
A or gsw was unstable. Here it looks like many
of the high light points were not stable, and it may be a good idea to
remove them before proceeding with the Ball-Berry fitting.
Light-Response Curves
# Make a plot to check light-response curves
xyplot(
A ~ Qin | curve_identifier,
data = licor_data$main_data,
type = 'b',
pch = 16,
auto = TRUE,
grid = TRUE,
ylim = c(-5, 50),
xlim = c(0, 2200),
xlab = paste0('Incident PPFD (', licor_data$units$Qin, ')'),
ylab = paste0('Net assimilation (', licor_data$units$A, ')')
)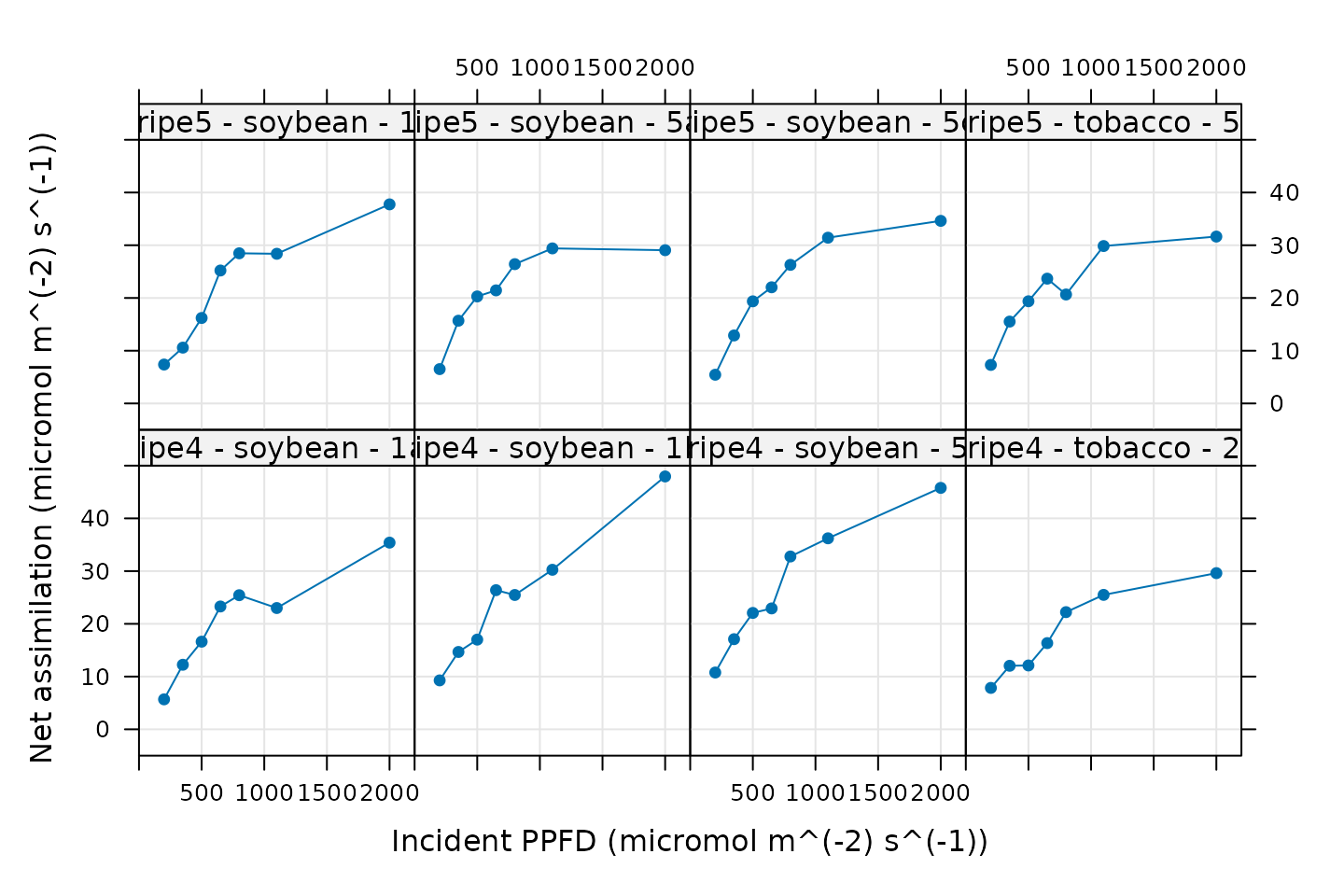
Here we are simply looking for a reasonable light-response curve
shape, where assimilation is low when light is low, assimilation has a
roughly linear response to initial increases in light intensity, and
then finally reaches a plateau. Any strong deviations from the expected
shape may indicate that the plant was stressed or otherwise behaving
abnormally, and we might not want to use such a curve for Ball-Berry
analysis. In this plot, three of the curves do not look like normal
C3 light response curves: ripe4 - soybean - 1a,
ripe4 - soybean - 1b, and ripe4 - soybean - 5.
These strange curves are likely a byproduct of the noise that was
intentionally added to the true measured data (see The Data). Nevertheless, it may be a good idea to
remove them before proceeding with the Ball-Berry fitting.
Cleaning the Licor Data
While checking over the plots in Qualitative Checks, two issues were
noticed: (1) some points were logged before stability was achieved and
(2) some of the curves look abnormal. In this section, we will
demonstrate how to remove the unstable points and the weird curves using
the remove_points function from PhotoGEA.
The following command will remove points where Stable is
0 or 1, keeping only the points where Stable is 2; this
condition means that all of the stability criteria were satisfied.
# Only keep points where stability was achieved
licor_data <- remove_points(
licor_data,
list(Stable = c(0, 1)),
method = 'exclude'
)Since we have identified a few curves that may not be acceptable for
Ball-Berry fitting, we can remove them via the
remove_points function from PhotoGEA:
The following command will remove all points from the curves that are not acceptable for Ball-Berry fitting:
# Define a list of curves to remove from the data set
curves_to_remove <- c(
'ripe4 - soybean - 1a',
'ripe4 - soybean - 1b',
'ripe4 - soybean - 5'
)
# Remove them
licor_data <- remove_points(
licor_data,
list(curve_identifier = curves_to_remove),
method = 'remove'
)Fitting Licor Data
Now that we have checked the data quality, we are ready to perform
the fitting. As they are produced by the instruments, Licor data files
do not include values of the Ball-Berry index; in fact, they do not even
include values of
and
required to calculate the Ball-Berry index. However, the
PhotoGEA package includes three functions to help with
these calculations: calculate_total_pressure,
calculate_gas_properties, and
calculate_ball_berry_index. Each of these requires an
exdf object containing Licor data. The units for each
required column will be checked in an attempt to avoid unit-related
errors. More information about these functions can be obtained from the
built-in help system by typing ?calculate_total_pressure,
?calculate_gas_properties, or
?calculate_ball_berry_index. Here we will use them
sequentially to calculate values of the Ball-Berry index:
# Calculate the total pressure in the Licor chamber
licor_data <- calculate_total_pressure(licor_data)
# Calculate additional gas properties, including `RHleaf` and `Csurface`
licor_data <- calculate_gas_properties(licor_data)
# Calculate the Ball-Berry index
licor_data <- calculate_ball_berry_index(licor_data)Together, these functions add several new columns to
licor_data, including one called bb_index,
which includes values of the Ball-Berry index. With this information, we
are now ready to perform the fitting procedure. For this operation, we
can use the fit_ball_berry function from the
PhotoGEA package, which fits a single Ball-Berry curve to
extract the Ball-Berry parameters. To apply this function to each curve
in a larger data set and then consolidate the results, we can use it in
conjunction with by and consolidate, which are
also part of PhotoGEA. (For more information about these
functions, see the built-in help menu entries by typing
?fit_ball_berry, ?by.exdf, or
?consolidate, or check out the Common Patterns
section of the Working
With Extended Data Frames vignette.) Together, these functions will
split apart the main data using the curve identifier column we defined
before (Basic Checks), make a linear fit of
against the Ball-Berry index, and return the resulting parameters and
fits:
# Fit a linear model to the Ball-Berry data
ball_berry_results <- consolidate(by(
licor_data, # The `exdf` object containing the curves
licor_data[, 'curve_identifier'], # A factor used to split `licor_data` into chunks
fit_ball_berry # The function to apply to each chunk of `licor_data`
))Viewing the Fitted Curves
Having made the fits, it is now a good idea to visually check them,
making sure they look reasonable. For this, we can use the
plot_ball_berry_fits function from PhotoGEA,
which will also show the points that were excluded from the fits during
the data cleaning step:
# Plot the Ball-Berry fits
plot_ball_berry_fit(ball_berry_results, 'curve_identifier')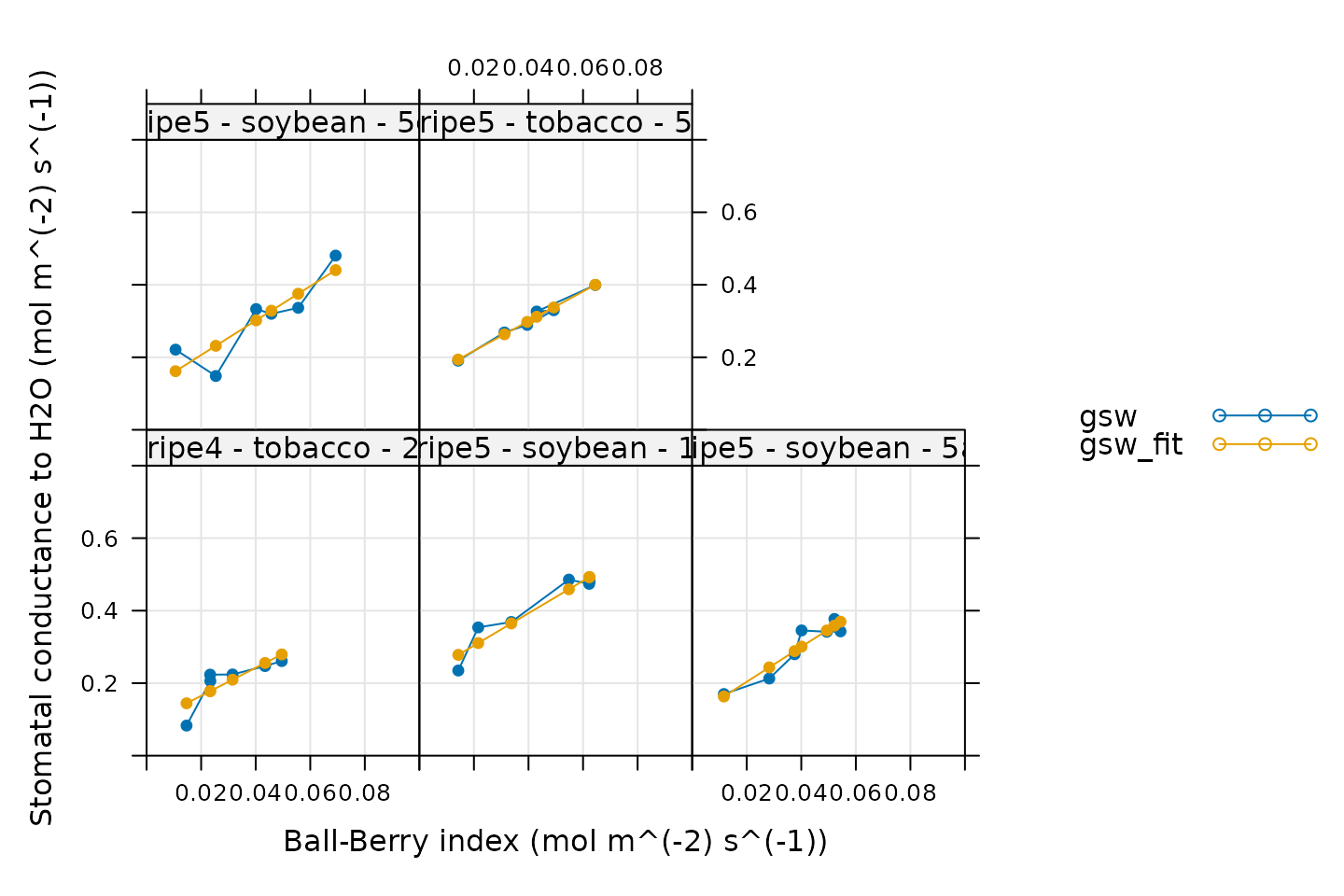
All these fits look good!
Viewing the Fitted Parameter Values
We can also take a look at the fitted Ball-Berry parameter values,
which are stored in ball_berry_results$parameters, another
exdf object. This object includes many columns but we only
care about a few of them. We can view them as follows:
# View the Ball-Berry parameters
columns_for_viewing <-
c('instrument', 'species', 'plot', 'bb_intercept', 'bb_slope', 'r_squared')
ball_berry_parameters <-
ball_berry_results$parameters[ , columns_for_viewing, TRUE]
print(ball_berry_parameters)
#>
#> Converting an `exdf` object to a `data.frame` before printing
#>
#> instrument [UserDefCon] (NA) species [UserDefCon] (NA) plot [UserDefCon] (NA)
#> 1 ripe4 tobacco 2
#> 2 ripe5 soybean 1
#> 3 ripe5 soybean 5a
#> 4 ripe5 soybean 5c
#> 5 ripe5 tobacco 5
#> bb_intercept [fit_ball_berry] (mol m^(-2) s^(-1))
#> 1 0.08778255
#> 2 0.21437406
#> 3 0.10722148
#> 4 0.11118324
#> 5 0.13565311
#> bb_slope [fit_ball_berry] (dimensionless) r_squared [fit_ball_berry] ()
#> 1 3.864316 0.6452714
#> 2 4.462244 0.9004803
#> 3 4.823671 0.8866346
#> 4 4.750743 0.7726284
#> 5 4.097481 0.9849869Extracting Average Values
Finally, we can extract average values of the Ball-Berry parameters
for each species using the basic_stats function from
PhotoGEA:
# Compute the average and standard error for the Ball-Berry slope and intercept
# for each species
ball_berry_averages <- basic_stats(
ball_berry_results$parameters,
'species'
)
# View the averages and errors
columns_to_view <- c(
'species',
'bb_intercept_avg', 'bb_intercept_stderr',
'bb_slope_avg', 'bb_slope_stderr'
)
print(ball_berry_averages[ , columns_to_view, TRUE])
#>
#> Converting an `exdf` object to a `data.frame` before printing
#>
#> species [UserDefCon] (NA)
#> 1 soybean
#> 2 tobacco
#> bb_intercept_avg [fit_ball_berry] (mol m^(-2) s^(-1))
#> 1 0.1442596
#> 2 0.1117178
#> bb_intercept_stderr [fit_ball_berry] (mol m^(-2) s^(-1))
#> 1 0.03507588
#> 2 0.02393528
#> bb_slope_avg [fit_ball_berry] (dimensionless)
#> 1 4.678886
#> 2 3.980898
#> bb_slope_stderr [fit_ball_berry] (dimensionless)
#> 1 0.1103477
#> 2 0.1165826Customizing Your Script
Note that most of the commands in this vignette have been written in a general way so they can be used as the basis for your own Ball-Berry analysis script (see Commands From This Document). In order to use them in your own script, some or all of the following changes may be required. There may also be others not specifically mentioned here.
Input Files
The file paths specified in file_paths will need to be
modified so they point to your Licor files. One way to do this in your
own script is to simply write out relative or absolute paths to the
files you wish to load. For example, you could replace the previous
definition of file_paths with this one:
# Define a vector of paths to the files we wish to load
file_paths <- c(
'myfile1.xlsx', # `myfile1.xlsx` must be in the current working directory
'C:/documents/myfile2' # This is an absolute path to `myfile2`
)You may also want to consider using the
choose_input_licor_files function from
PhotoGEA; this function will create a pop-up browser window
where you can interactively select a set of files. Sometimes this is
more convenient than writing out file paths or names. For example, you
could replace the previous definition of file_paths with
this one:
# Interactively define a vector of paths to the files we wish to load
file_paths <- choose_input_licor_files()Unfortunately, choose_input_licor_files is only
available in interactive R sessions running on Microsoft Windows, but
there is also a platform-independent option:
choose_input_files. See the Translation section of
the Developing a
Data Analysis Pipeline vignette for more details.
Curve Identifier
Depending on which user constants are defined in your Licor Excel
files, you may need to modify the definition of the
curve_identifier column.
Excluded Curves
Depending on the qualitative data checks, you may need to change the
definition of the curves_to_remove vector.
Averages and Standard Errors
Depending on how your data is organized, you may want to change the column used to divide the data when calculating averages and standard errors.
Plots
You may need to change the axis limits in some or all of the plots.
Alternatively, you can remove them, allowing xyplot to
automatically choose them for you.
Saving Results
You may want to use write.csv to save some or all of the
fitting results as csv files. For example, the following
commands will allow you to interactively choose output filenames for the
resulting csv files:
write.csv(ball_berry_results$fits, file.choose(), row.names = FALSE)
write.csv(ball_berry_results$parameters, file.choose(), row.names = FALSE)
write.csv(ball_berry_averages, file.choose(), row.names = FALSE)Commands From This Document
The following code chunk includes all the central commands used throughout this document. They are compiled here to make them easy to copy/paste into a text file to initialize your own script. Annotation has also been added to clearly indicate the four steps involved in data analysis, as described in the Developing a Data Analysis Pipeline vignette.
###
### PRELIMINARIES:
### Loading packages, defining constants, creating helping functions, etc.
###
# Load required packages
library(PhotoGEA)
library(lattice)
###
### TRANSLATION:
### Creating convenient R objects from raw data files
###
## IMPORTANT: When loading your own files, it is not advised to use
## `PhotoGEA_example_file_path` as in the code below. Instead, write out the
## names or use the `choose_input_licor_files` function.
# Define a vector of paths to the files we wish to load; in this case, we are
# loading example files included with the PhotoGEA package
file_paths <- c(
PhotoGEA_example_file_path('ball_berry_1.xlsx'),
PhotoGEA_example_file_path('ball_berry_2.xlsx')
)
# Load each file, storing the result in a list
licor_exdf_list <- lapply(file_paths, function(fpath) {
read_gasex_file(fpath, 'time')
})
# Get the names of all columns that are present in all of the Licor files
columns_to_keep <- do.call(identify_common_columns, licor_exdf_list)
# Extract just these columns
licor_exdf_list <- lapply(licor_exdf_list, function(x) {
x[ , columns_to_keep, TRUE]
})
# Use `rbind` to combine all the data
licor_data <- do.call(rbind, licor_exdf_list)
###
### VALIDATION:
### Organizing the data, checking its consistency and quality, cleaning it
###
# Create a new identifier column formatted like `instrument - species - plot`
licor_data[ , 'curve_identifier'] <-
paste(licor_data[ , 'instrument'], '-', licor_data[ , 'species'], '-', licor_data[ , 'plot'])
# Make sure the data meets basic requirements
check_response_curve_data(licor_data, 'curve_identifier', 7, 'Qin', 1.0)
# Make a plot to check humidity control
xyplot(
RHcham + `Humidifier_%` + `Desiccant_%` ~ Qin | curve_identifier,
data = licor_data$main_data,
type = 'b',
pch = 16,
auto = TRUE,
grid = TRUE,
ylim = c(0, 100),
xlim = c(0, 2200),
xlab = paste0('Incident PPFD (', licor_data$units$Qin, ')')
)
# Make a plot to check temperature control
xyplot(
TleafCnd + Txchg ~ Qin | curve_identifier,
data = licor_data$main_data,
type = 'b',
pch = 16,
auto = TRUE,
grid = TRUE,
ylim = c(25, 40),
xlim = c(0, 2200),
xlab = paste0('Incident PPFD (', licor_data$units$Qin, ')'),
ylab = paste0('Temperature (', licor_data$units$TleafCnd, ')')
)
# Make a plot to check CO2 control
xyplot(
CO2_s + CO2_r ~ Qin | curve_identifier,
data = licor_data$main_data,
type = 'b',
pch = 16,
auto = TRUE,
grid = TRUE,
ylim = c(360, 480),
xlim = c(0, 2200),
xlab = paste0('Incident PPFD (', licor_data$units$Qin, ')'),
ylab = paste0('CO2 concentration (', licor_data$units$CO2_r, ')')
)
xyplot(
`A:OK` + `gsw:OK` + Stable ~ Qin | curve_identifier,
data = licor_data$main_data,
type = 'b',
pch = 16,
auto = TRUE,
grid = TRUE,
xlim = c(0, 2200),
xlab = paste0('Incident PPFD (', licor_data$units$Qin, ')')
)
# Make a plot to check light-response curves
xyplot(
A ~ Qin | curve_identifier,
data = licor_data$main_data,
type = 'b',
pch = 16,
auto = TRUE,
grid = TRUE,
ylim = c(-5, 50),
xlim = c(0, 2200),
xlab = paste0('Incident PPFD (', licor_data$units$Qin, ')'),
ylab = paste0('Net assimilation (', licor_data$units$A, ')')
)
## IMPORTANT: When analyzing your own files, it is not advised to remove any
## points for the initial fits. Only remove unstable or unusual points if it is
## necessary in order to get good fits.
# Only keep points where stability was achieved
licor_data <- remove_points(
licor_data,
list(Stable = c(0, 1)),
method = 'exclude'
)
# Define a list of curves to remove from the data set
curves_to_remove <- c(
'ripe4 - soybean - 1a',
'ripe4 - soybean - 1b',
'ripe4 - soybean - 5'
)
# Remove them
licor_data <- remove_points(
licor_data,
list(curve_identifier = curves_to_remove),
method = 'remove'
)
###
### PROCESSING:
### Extracting new pieces of information from the data
###
# Calculate the total pressure in the Licor chamber
licor_data <- calculate_total_pressure(licor_data)
# Calculate additional gas properties, including `RHleaf` and `Csurface`
licor_data <- calculate_gas_properties(licor_data)
# Calculate the Ball-Berry index
licor_data <- calculate_ball_berry_index(licor_data)
# Fit a linear model to the Ball-Berry data
ball_berry_results <- consolidate(by(
licor_data, # The `exdf` object containing the curves
licor_data[, 'curve_identifier'], # A factor used to split `licor_data` into chunks
fit_ball_berry # The function to apply to each chunk of `licor_data`
))
# Plot the Ball-Berry fits
plot_ball_berry_fit(ball_berry_results, 'curve_identifier')
# View the Ball-Berry parameters
columns_for_viewing <-
c('instrument', 'species', 'plot', 'bb_intercept', 'bb_slope', 'r_squared')
ball_berry_parameters <-
ball_berry_results$parameters[ , columns_for_viewing, TRUE]
print(ball_berry_parameters)
###
### SYNTHESIS:
### Using plots and statistics to help draw conclusions from the data
###
# Compute the average and standard error for the Ball-Berry slope and intercept
# for each species
ball_berry_averages <- basic_stats(
ball_berry_results$parameters,
'species'
)
# View the averages and errors
columns_to_view <- c(
'species',
'bb_intercept_avg', 'bb_intercept_stderr',
'bb_slope_avg', 'bb_slope_stderr'
)
print(ball_berry_averages[ , columns_to_view, TRUE])