Fits a C3 assimilation model to an A-Ci curve
fit_c3_aci.RdFits the Farquhar-von-Caemmerer-Berry model to an experimentally measured C3 A-Ci curve.
It is possible to fit the following parameters: alpha_g,
alpha_old, alpha_s, alpha_t, Gamma_star_at_25,
gmc_at_25, J_at_25, Kc_at_25, Ko_at_25,
RL_at_25, Tp_at_25, and Vcmax_at_25.
By default, only a subset of these parameters are actually fit:
alpha_old, J_at_25, RL_at_25, Tp_at_25, and
Vcmax_at_25. This can be altered using the fit_options argument,
as described below.
Best-fit parameters are found using maximum likelihood fitting, where the
optimizer (optim_fun) is used to minimize the error function (defined
by error_function_c3_aci).
Once best-fit parameters are found, confidence intervals are calculated
using confidence_intervals_c3_aci, and unreliable parameter
estimates are removed.
For temperature-dependent parameters, best-fit values and confidence intervals are returned at 25 degrees C and at leaf temperature.
See below for more details.
Usage
fit_c3_aci(
replicate_exdf,
Ca_atmospheric = NA,
a_column_name = 'A',
ca_column_name = 'Ca',
ci_column_name = 'Ci',
gamma_star_norm_column_name = 'Gamma_star_norm',
gmc_norm_column_name = 'gmc_norm',
j_norm_column_name = 'J_norm',
kc_norm_column_name = 'Kc_norm',
ko_norm_column_name = 'Ko_norm',
oxygen_column_name = 'Oxygen',
rl_norm_column_name = 'RL_norm',
total_pressure_column_name = 'total_pressure',
tp_norm_column_name = 'Tp_norm',
vcmax_norm_column_name = 'Vcmax_norm',
sd_A = 'RMSE',
Wj_coef_C = 4.0,
Wj_coef_Gamma_star = 8.0,
optim_fun = optimizer_deoptim(200),
lower = list(),
upper = list(),
fit_options = list(),
cj_crossover_min = NA,
cj_crossover_max = NA,
relative_likelihood_threshold = 0.147,
hard_constraints = 0,
calculate_confidence_intervals = TRUE,
remove_unreliable_param = 2,
debug_mode = FALSE,
...
)Arguments
- replicate_exdf
An
exdfobject representing one CO2 response curve.- Ca_atmospheric
The atmospheric CO2 concentration (with units of
micromol mol^(-1)); this will be used byestimate_operating_pointto estimate the operating point. A value ofNAdisables this feature.- a_column_name
The name of the column in
replicate_exdfthat contains the net assimilation inmicromol m^(-2) s^(-1).- ca_column_name
The name of the column in
replicate_exdfthat contains the ambient CO2 concentration inmicromol mol^(-1). If values ofCaare not available, they can be set toNA. In this case, it will not be possible to estimate the operating point, andapply_gmwill not be able to calculate the CO2 drawdown across the stomata.- ci_column_name
The name of the column in
replicate_exdfthat contains the intercellular CO2 concentration inmicromol mol^(-1).- gamma_star_norm_column_name
The name of the column in
replicate_exdfthat contains the normalizedGamma_starvalues (with units ofnormalized to Gamma_star at 25 degrees C).- gmc_norm_column_name
The name of the column in
replicate_exdfthat contains the normalized mesophyll conductance values (with units ofnormalized to gmc at 25 degrees C).- j_norm_column_name
The name of the column in
replicate_exdfthat contains the normalizedJvalues (with units ofnormalized to J at 25 degrees C).- kc_norm_column_name
The name of the column in
replicate_exdfthat contains the normalizedKcvalues (with units ofnormalized to Kc at 25 degrees C).- ko_norm_column_name
The name of the column in
replicate_exdfthat contains the normalizedKovalues (with units ofnormalized to Ko at 25 degrees C).- oxygen_column_name
The name of the column in
replicate_exdfthat contains the concentration of O2 in the ambient air, expressed as a percentage (commonly 21% or 2%); the units must bepercent.- rl_norm_column_name
The name of the column in
replicate_exdfthat contains the normalizedRLvalues (with units ofnormalized to RL at 25 degrees C).- total_pressure_column_name
The name of the column in
replicate_exdfthat contains the total pressure inbar.- tp_norm_column_name
The name of the column in
replicate_exdfthat contains the normalizedTpvalues (with units ofnormalized to Tp at 25 degrees C).- vcmax_norm_column_name
The name of the column in
replicate_exdfthat contains the normalizedVcmaxvalues (with units ofnormalized to Vcmax at 25 degrees C).- sd_A
A value of the standard deviation of measured
Avalues, or the name of a method for determining the deviation; currently, the only supported option is'RMSE'.- Wj_coef_C
A coefficient in the equation for RuBP-regeneration-limited carboxylation, whose value depends on assumptions about the NADPH and ATP requirements of RuBP regeneration; see
calculate_c3_assimilationfor more information.- Wj_coef_Gamma_star
A coefficient in the equation for RuBP-regeneration-limited carboxylation, whose value depends on assumptions about the NADPH and ATP requirements of RuBP regeneration; see
calculate_c3_assimilationfor more information.- optim_fun
An optimization function that accepts the following input arguments: an initial guess, an error function, lower bounds, and upper bounds. It should return a list with the following elements:
par,convergence,feval, andconvergence_msg. Seeoptimizersfor a list of available options.- lower
A list of named numeric elements representing lower bounds to use when fitting. Values supplied here override the default values (see details below). For example,
lower = list(Vcmax_at_25 = 10)sets the lower limit forVcmax_at_25to 10 micromol / m^2 / s.- upper
A list of named numeric elements representing upper bounds to use when fitting. Values supplied here override the default values (see details below). For example,
upper = list(Vcmax_at_25 = 200)sets the upper limit forVcmax_at_25to 200 micromol / m^2 / s.- fit_options
A list of named elements representing fit options to use for each parameter. Values supplied here override the default values (see details below). Each element must be
'fit','column', or a numeric value. A value of'fit'means that the parameter will be fit; a value of'column'means that the value of the parameter will be taken from a column inreplicate_exdfof the same name; and a numeric value means that the parameter will be set to that value. For example,fit_options = list(alpha_g = 0, Vcmax_at_25 = 'fit', Tp_at_25 = 'column')means thatalpha_gwill be set to 0,Vcmax_at_25will be fit, andTp_at_25will be set to the values in theTp_at_25column ofreplicate_exdf.- cj_crossover_min
The minimum value of
Cc(in ppm) whereAjis allowed to become the overall rate-limiting factor. Ifcj_crossover_minis set toNA, this restriction will not be applied.- cj_crossover_max
The maximim value of
Cc(in ppm) whereWjis allowed to be smaller thanWc. Ifcj_crossover_maxis set toNA, this restriction will not be applied.- relative_likelihood_threshold
To be passed to
confidence_intervals_c3_aciwhencalculate_confidence_intervalsisTRUE.- hard_constraints
To be passed to
calculate_c3_assimilation; see that function for more details.- calculate_confidence_intervals
A logical value indicating whether or not to estimate confidence intervals for the fitting parameters using
confidence_intervals_c3_aci.- remove_unreliable_param
An integer value indicating the rules to use when identifying and removing unreliable parameter estimates. A value of 2 is the most conservative option. A value of 0 disables this feature, which is not typically recommended. It is also possible to directly specify the trust values to remove; for example,
'unreliable (process never limiting)'is equivalent to 1. See below for more details.- debug_mode
A logical (
TRUEorFALSE) variable indicating whether to operate in debug mode. In debug mode, information aboutreplicate_exdf, the initial guess, each guess supplied from the optimizer, and the final outcome is printed; this can be helpful when troubleshooting issues with a particular curve.- ...
Additional arguments to be passed to
calculate_c3_assimilation.
Details
This function calls apply_gm and
calculate_c3_assimilation to calculate values of net
assimilation. The user-supplied optimization function is used to vary the
values of alpha_g, alpha_old, alpha_s, alpha_t,
Gamma_star_at_25, gmc_at_25, J_at_25, Kc_at_25,
Ko_at_25, RL_at_25, Tp_at_25, and Vcmax_at_25 to
find ones that best reproduce the experimentally measured values of net
assimilation. By default, the following options are used for the fits:
alpha_g: lower = 0, upper = 10, fit_option = 0alpha_old: lower = 0, upper = 10, fit_option ='fit'alpha_s: lower = 0, upper = 10, fit_option = 0alpha_t: lower = 0, upper = 10, fit_option = 0Gamma_star_at_25: lower = -20, upper = 200, fit_option ='column'gmc_at_25: lower = -1, upper = 10, fit_option =InfJ_at_25: lower = -50, upper = 1000, fit_option ='fit'Kc_at_25: lower = -50, upper = 1000, fit_option ='column'Ko_at_25: lower = -50, upper = 1000, fit_option ='column'RL_at_25: lower = -10, upper = 100, fit_option ='fit'Tp_at_25: lower = -10, upper = 100, fit_option ='fit'Vcmax_at_25: lower = -50, upper = 1000, fit_option ='fit'
With these settings, the "new" alpha parameters are set to 0; values of
Gamma_star_at_25, Kc_at_25, and Ko_at_25 are taken from
the Gamma_star_at_25, Kc_at_25, and Ko_at_25 columns of
replicate_exdf; mesophyll conductance (gmc_at_25) is set to
inifinity (so Cc = Ci); and the other parameters are fit during the
process (see fit_options above). The bounds are chosen liberally to
avoid any bias.
An initial guess for the parameters is generated by calling
initial_guess_c3_aci as follows:
cc_threshold_rlis set to 100 micromol / mol.If
alpha_gis being fit, thealpha_gargument ofinitial_guess_c3_aciis set to 0.5; otherwise, the argument is set to the value specified by the fit options.If
alpha_oldis being fit, thealpha_oldargument ofinitial_guess_c3_aciis set to 0.5; otherwise, the argument is set to the value specified by the fit options.if
alpha_sis being fit, thealpha_sargument ofinitial_guess_c3_aciis set to0.3 * (1 - alpha_g); otherwise, the argument is set to the value specified by the fit options.if
alpha_tis being fit, thealpha_targument ofinitial_guess_c3_aciis set to 0; otherwise, the argument is set to the value specified by the fit options.If
Gamma_star_at_25is being fit, theGamma_star_at_25argument ofinitial_guess_c3_aciis set to 40; otherwise, the argument is set to the value specified by the fit options.If
gmc_at_25is being fit, thegmc_at_25argument ofinitial_guess_c3_aciis set to 1; otherwise, the argument is set to the value specified by the fit options.If
Kc_at_25is being fit, theKc_at_25argument ofinitial_guess_c3_aciis set to 400; otherwise, the argument is set to the value specified by the fit options.If
Ko_at_25is being fit, theKo_at_25argument ofinitial_guess_c3_aciis set to 275; otherwise, the argument is set to the value specified by the fit options.
Note that any fixed values specified in the fit options will override the values returned by the guessing function.
The fit is made by creating an error function using
error_function_c3_aci and minimizing its value using
optim_fun, starting from the initial guess described above. The
optimizer_deoptim optimizer is used by default since it has been
found to reliably return great fits. However, it is a slow optimizer. If speed
is important, consider reducing the number of generations or using
optimizer_nmkb, but be aware that this optimizer is more likely
to get stuck in a local minimum.
The photosynthesis model represented by calculate_c3_assimilation is
not smooth in the sense that small changes in the input parameters do not
necessarily cause changes in its outputs. This is related to the final step in
the calculations, where the overall assimilation rate is taken to be the
minimum of three enzyme-limited rates. For example, if the assimilation rate
is never TPU-limited, modifying Tp_at_25 will not change the model's
outputs. For this reason, derivative-based optimizers tend to struggle when
fitting C3 A-Ci curves. Best results are obtained using derivative-free
methods.
Sometimes the optimizer may choose a set of parameter values where one or more
of the potential limiting carboxylation rates (Wc, Wj, or
Wp) is never the smallest rate. In this case, the corresponding
parameter estimates (Vcmax, J, or alpha_old & Tp)
will be severely unreliable. This will be indicated by a value of
'unreliable (process never limiting)' in the corresponding trust column
(for example, Vcmax_trust). If remove_unreliable_param is
1 or larger, then such parameter estimates (and the corresponding
rates) will be replaced by NA in the fitting results.
It is also possible that the upper limit of the confidence interval for a
parameter is infinity; this indicates a potentially unreliable parameter
estimate. This will be indicated by a value of
'unreliable (infinite upper limit)' in the corresponding trust column
(for example, Vcmax_trust). If remove_unreliable_param is
2 or larger, then such parameter estimates (but not the corresponding
rates) will be replaced by NA in the fitting results.
The trust value for fully reliable parameter estimates is set to
'reliable' and they will never be replaced by NA.
Once the best-fit parameters have been determined, this function also
estimates the operating value of Cc from the atmospheric CO2
concentration atmospheric_ca using
estimate_operating_point, and then uses that value to estimate
the modeled An at the operating point via
calculate_c3_assimilation. It also estimates the
Akaike information criterion (AIC).
This function assumes that replicate_exdf represents a single
C3 A-Ci curve. To fit multiple curves at once, this function is often used
along with by.exdf and consolidate.
Value
A list with three elements:
fits: Anexdfobject including the original contents ofreplicate_exdfalong with several new columns:The fitted values of net assimilation will be stored in a column whose name is determined by appending
'_fit'to the end ofa_column_name; typically, this will be'A_fit'.Residuals (measured - fitted) will be stored in a column whose name is determined by appending
'_residuals'to the end ofa_column_name; typically, this will be'A_residuals'.Values of fitting parameters at 25 degrees C will be stored in the
Gamma_star_at_25,gmc_at_25,J_at_25,Kc_at_25,Ko_at_25,RL_at_25,Tp_at_25, andVcmax_at_25columns.The other outputs from
calculate_c3_assimilationwill be stored in columns with the usual names:alpha_g,alpha_old,alpha_s,alpha_t,Gamma_star_tl,gmc_tl,Kc_tl,Ko_tl,Tp_tl,Vcmax_tl,RL_tl,J_tl,Wc,Wj,Wp,Vc,Ac,Aj, andAp.
fits_interpolated: Anexdfobject including the calculated assimilation rates at a fine spacing ofCivalues (step size of 1micromol mol^(-1)).parameters: Anexdfobject including the identifiers, fitting parameters, and convergence information for the A-Ci curve:The number of points where
An = Ac,An = Aj, andAn = Apare stored in then_Ac_limiting,n_Aj_limiting, andn_Ap_limitingcolumns.The best-fit values are stored in the
alpha_g,alpha_old,alpha_s,alpha_t,Gamma_star_at_25,gmc_at_25,J_at_25,Kc_at_25,Ko_at_25,RL_at_25,Tp_at_25, andVcmax_at_25columns. Ifcalculate_confidence_intervalsisTRUE, upper and lower limits for each of these parameters will also be included.For parameters that depend on leaf temperature, the average leaf-temperature-dependent values are stored in
Gamma_star_tl_avg,gmc_tl_avg,J_tl_avg,Kc_tl_avg,Ko_tl_avg,RL_tl_avg,Tp_tl_avg, andVcmax_tl_avg.Information about the operating point is stored in
operating_Cc,operating_Ci,operating_An, andoperating_An_model.The
convergencecolumn indicates whether the fit was successful (==0) or if the optimizer encountered a problem (!=0).The
fevalcolumn indicates how many cost function evaluations were required while finding the optimal parameter values.The residual stats as returned by
residual_statsare included as columns with the default names:dof,RSS,RMSE, etc.The Akaike information criterion is included in the
AICcolumn.
Examples
# Read an example Licor file included in the PhotoGEA package
licor_file <- read_gasex_file(
PhotoGEA_example_file_path('c3_aci_1.xlsx')
)
# Define a new column that uniquely identifies each curve
licor_file[, 'species_plot'] <-
paste(licor_file[, 'species'], '-', licor_file[, 'plot'] )
# Organize the data
licor_file <- organize_response_curve_data(
licor_file,
'species_plot',
c(9, 10, 16),
'CO2_r_sp'
)
# Calculate the total pressure in the Licor chamber
licor_file <- calculate_total_pressure(licor_file)
# Calculate temperature-dependent values of C3 photosynthetic parameters
licor_file <- calculate_temperature_response(licor_file, c3_temperature_param_bernacchi)
# For these examples, we will use a faster (but sometimes less reliable)
# optimizer so they run faster
optimizer <- optimizer_nmkb(1e-7)
# We can fit just one curve from the data set, although it is rare to do this
one_result <- fit_c3_aci(
licor_file[licor_file[, 'species_plot'] == 'tobacco - 1', , TRUE],
Ca_atmospheric = 420,
optim_fun = optimizer
)
# We can fit the same curve, but allow alpha_old and Gamma_star_at_25 to vary
one_result_v2 <- fit_c3_aci(
licor_file[licor_file[, 'species_plot'] == 'tobacco - 1', , TRUE],
Ca_atmospheric = 420,
fit_options = list(Gamma_star_at_25 = 'fit', alpha_old = 'fit'),
optim_fun = optimizer
)
# Fit all curves in the data set (it is more common to do this)
aci_results <- consolidate(by(
licor_file,
licor_file[, 'species_plot'],
fit_c3_aci,
Ca_atmospheric = 420,
optim_fun = optimizer
))
# View the fitting parameters for each species / plot
col_to_keep <- c(
'species', 'plot', # identifiers
'n_Ac_limiting', 'n_Aj_limiting', 'n_Ap_limiting', # number of points where
# each process is limiting
'Tp_at_25', 'J_at_25', 'RL_at_25', 'Vcmax_at_25', # parameters scaled to 25 degrees C
'J_tl_avg', 'RL_tl_avg', 'Vcmax_tl_avg', # average temperature-dependent values
'operating_Ci', 'operating_An', 'operating_An_model', # operating point info
'dof', 'RSS', 'MSE', 'RMSE', 'RSE', # residual stats
'convergence', 'convergence_msg', 'feval', 'optimum_val' # convergence info
)
aci_results$parameters[ , col_to_keep, TRUE]
#>
#> Converting an `exdf` object to a `data.frame` before printing
#>
#> species [UserDefCon] (NA) plot [UserDefCon] (NA)
#> 1 soybean 5a
#> 2 tobacco 1
#> 3 tobacco 2
#> n_Ac_limiting [identify_c3_unreliable_points] ()
#> 1 8
#> 2 8
#> 3 9
#> n_Aj_limiting [identify_c3_unreliable_points] ()
#> 1 5
#> 2 5
#> 3 4
#> n_Ap_limiting [identify_c3_unreliable_points] ()
#> 1 0
#> 2 0
#> 3 0
#> Tp_at_25 [fit_c3_aci] (micromol m^(-2) s^(-1))
#> 1 NA
#> 2 NA
#> 3 NA
#> J_at_25 [fit_c3_aci] (micromol m^(-2) s^(-1))
#> 1 190.8859
#> 2 231.7749
#> 3 206.4820
#> RL_at_25 [fit_c3_aci] (micromol m^(-2) s^(-1))
#> 1 0.5770711
#> 2 0.8490820
#> 3 0.3815406
#> Vcmax_at_25 [fit_c3_aci] (micromol m^(-2) s^(-1))
#> 1 144.5530
#> 2 157.7597
#> 3 124.2033
#> J_tl_avg [fit_c3_aci] (micromol m^(-2) s^(-1))
#> 1 268.1496
#> 2 319.2523
#> 3 285.9199
#> RL_tl_avg [fit_c3_aci] (micromol m^(-2) s^(-1))
#> 1 0.8149562
#> 2 1.1742260
#> 3 0.5306255
#> Vcmax_tl_avg [fit_c3_aci] (micromol m^(-2) s^(-1))
#> 1 232.0402
#> 2 245.8747
#> 3 195.1092
#> operating_Ci [estimate_operating_point] (micromol mol^(-1))
#> 1 264.3297
#> 2 294.7032
#> 3 301.2673
#> operating_An [estimate_operating_point] (micromol m^(-2) s^(-1))
#> 1 31.00316
#> 2 37.51608
#> 3 31.57904
#> operating_An_model [fit_c3_aci] (micromol m^(-2) s^(-1))
#> 1 33.29502
#> 2 40.46328
#> 3 33.05702
#> dof [residual_stats] (NA) RSS [residual_stats] ((micromol m^(-2) s^(-1))^2)
#> 1 8 149.01920
#> 2 8 53.22941
#> 3 8 49.78449
#> MSE [residual_stats] ((micromol m^(-2) s^(-1))^2)
#> 1 11.463015
#> 2 4.094570
#> 3 3.829576
#> RMSE [residual_stats] (micromol m^(-2) s^(-1))
#> 1 3.385707
#> 2 2.023504
#> 3 1.956930
#> RSE [residual_stats] (micromol m^(-2) s^(-1)) convergence [fit_c3_aci] ()
#> 1 4.315947 0
#> 2 2.579472 0
#> 3 2.494606 0
#> convergence_msg [fit_c3_aci] () feval [fit_c3_aci] ()
#> 1 Successful convergence 243
#> 2 Successful convergence 214
#> 3 Successful convergence 218
#> optimum_val [fit_c3_aci] ()
#> 1 34.30052
#> 2 27.60900
#> 3 27.17410
# View the fits for each species / plot
plot_c3_aci_fit(aci_results, 'species_plot', 'Ci')
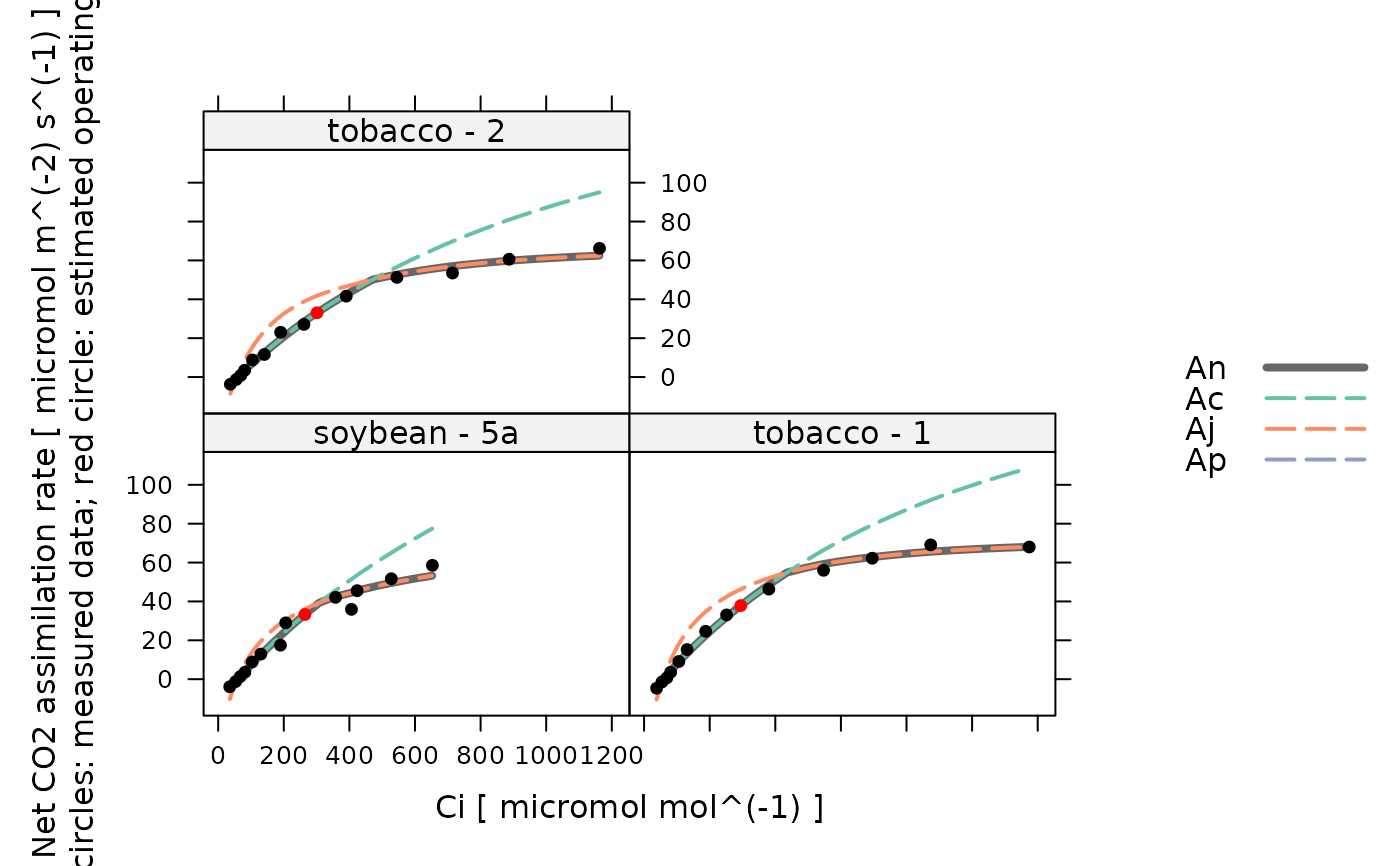 # View the residuals for each species / plot
lattice::xyplot(
A_residuals ~ Ci | species_plot,
data = aci_results$fits$main_data,
type = 'b',
pch = 16,
auto = TRUE,
grid = TRUE,
xlab = paste0('Intercellular CO2 concentration (', aci_results$fits$units$Ci, ')'),
ylab = paste0('Assimilation rate residuals (', aci_results$fits$units$A_residuals, ')')
)
# View the residuals for each species / plot
lattice::xyplot(
A_residuals ~ Ci | species_plot,
data = aci_results$fits$main_data,
type = 'b',
pch = 16,
auto = TRUE,
grid = TRUE,
xlab = paste0('Intercellular CO2 concentration (', aci_results$fits$units$Ci, ')'),
ylab = paste0('Assimilation rate residuals (', aci_results$fits$units$A_residuals, ')')
)
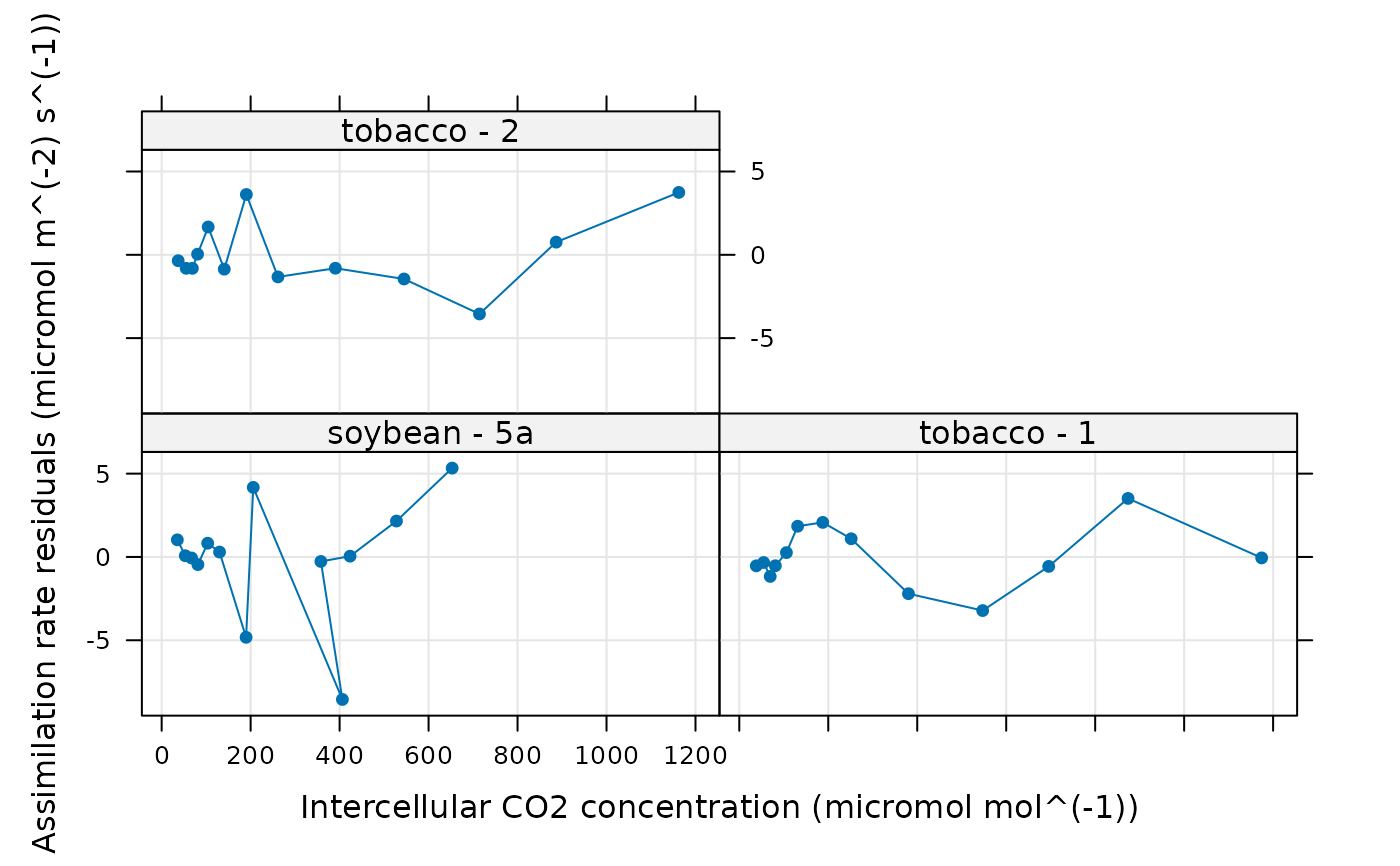 # In some of the curves above, there are no points where carboxylation is TPU
# limited. Estimates of Tp are therefore unreliable and are removed.
# In some of the curves above, there are no points where carboxylation is TPU
# limited. Estimates of Tp are therefore unreliable and are removed.