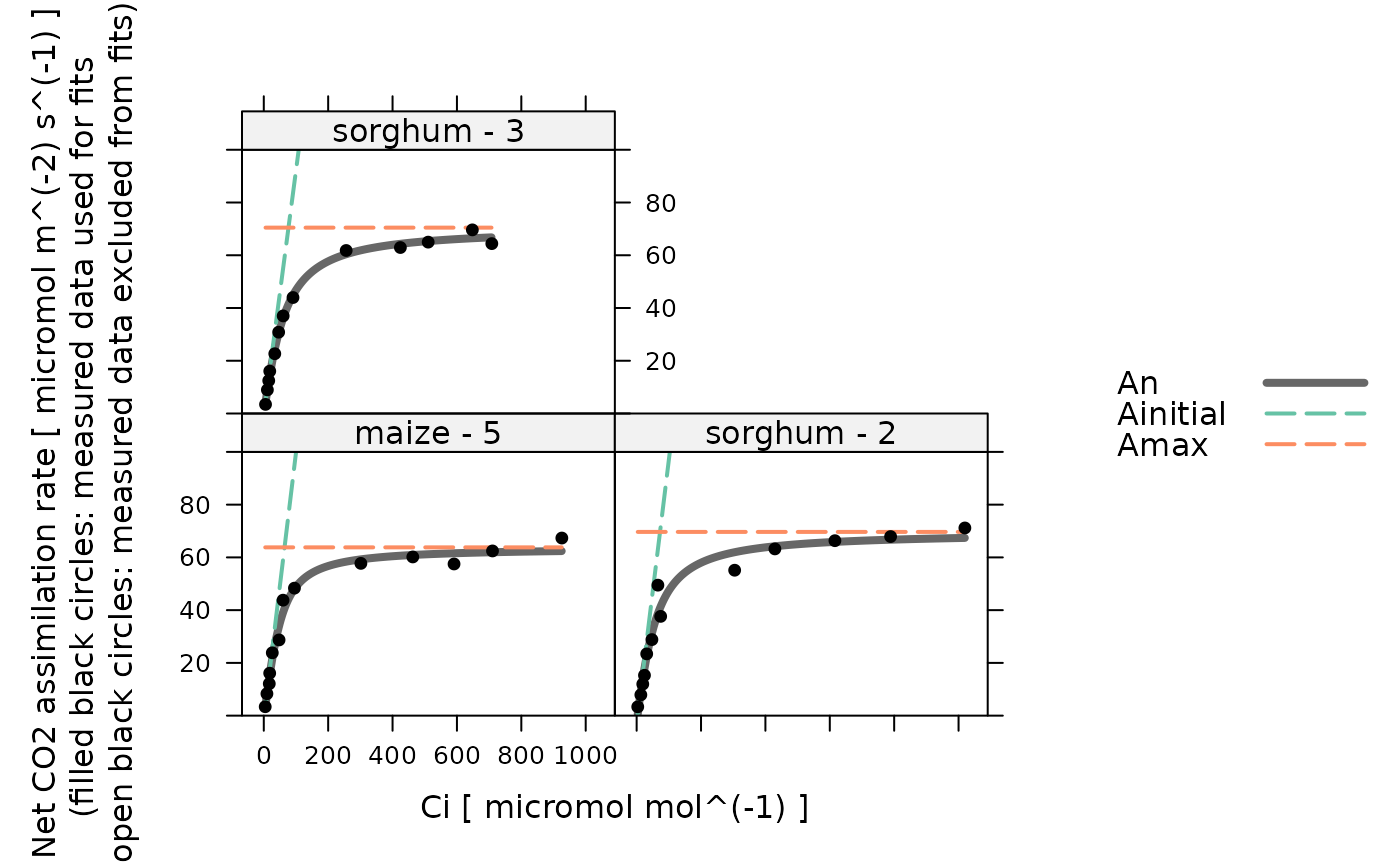
Plot the results of a hyperbolic C4 CO2 response curve fit
plot_c4_aci_hyperbola_fit.RdPlots the output from fit_c4_aci_hyperbola.
Usage
plot_c4_aci_hyperbola_fit(
fit_results,
identifier_column_name,
a_column_name = 'A',
ci_column_name = 'Ci',
...
)Arguments
- fit_results
A list of three
exdfobjects namedfits,parameters, andfits_interpolated, as calculated byfit_c4_aci_hyperbola.- identifier_column_name
The name of a column in each element of
fit_resultswhose value can be used to identify each response curve within the data set; often, this is'curve_identifier'.- a_column_name
The name of the columns in the elements of
fit_resultsthat contain the net assimilation inmicromol m^(-2) s^(-1); should be the same value that was passed tofit_c4_aci_hyperbola.- ci_column_name
The name of the columns in the elements of
fit_resultsthat contain the intercellular CO2 concentration inmicromol mol^(-1); should be the same value that was passed tofit_c4_aci_hyperbola.- ...
Additional arguments to be passed to
xyplot.
Details
This is a convenience function for plotting the results of a C4 A-Ci curve
fit. It is typically used for displaying several fits at once, in which case
fit_results is actually the output from calling
consolidate on a list created by calling by.exdf
with FUN = fit_c4_aci_hyperbola.
The resulting plot will show curves for the fitted rates An,
Ainitial, and Amax, along with points for the measured values of
A.
Internally, this function uses xyplot to perform the
plotting. Optionally, additional arguments can be passed to xyplot.
These should typically be limited to things like xlim, ylim,
main, and grid, since many other xyplot arguments will be
set internally (such as xlab, ylab, auto, and others).
See the help file for fit_c4_aci_hyperbola for an example using
this function.
Value
A trellis object created by lattice::xyplot.
Examples
# Read an example Licor file included in the PhotoGEA package
licor_file <- read_gasex_file(
PhotoGEA_example_file_path('c4_aci_1.xlsx')
)
# Define a new column that uniquely identifies each curve
licor_file[, 'species_plot'] <-
paste(licor_file[, 'species'], '-', licor_file[, 'plot'] )
# Organize the data
licor_file <- organize_response_curve_data(
licor_file,
'species_plot',
c(9, 10, 16),
'CO2_r_sp'
)
# Fit all curves in the data set
aci_results <- consolidate(by(
licor_file,
licor_file[, 'species_plot'],
fit_c4_aci_hyperbola
))
# View the fits for each species / plot
plot_c4_aci_hyperbola_fit(aci_results, 'species_plot', ylim = c(0, 100))