Generate an error function for C4 A-Ci curve fitting
error_function_c4_aci.RdCreates a function that returns an error value (the negative of the natural
logarithm of the likelihood) representing the amount of agreement between
modeled and measured An values. When this function is minimized, the
likelihood is maximized.
Internally, this function uses apply_gm to calculate Cc,
and then uses link{calculate_c4_assimilation} to calculate assimilation
rate values that are compared to the measured ones.
Usage
error_function_c4_aci(
replicate_exdf,
fit_options = list(),
sd_A = 1,
x_etr = 0.4,
a_column_name = 'A',
ao_column_name = 'ao',
ci_column_name = 'Ci',
gamma_star_column_name = 'gamma_star',
gmc_norm_column_name = 'gmc_norm',
j_norm_column_name = 'J_norm',
kc_column_name = 'Kc',
ko_column_name = 'Ko',
kp_column_name = 'Kp',
oxygen_column_name = 'Oxygen',
rl_norm_column_name = 'RL_norm',
total_pressure_column_name = 'total_pressure',
vcmax_norm_column_name = 'Vcmax_norm',
vpmax_norm_column_name = 'Vpmax_norm',
hard_constraints = 0,
debug_mode = FALSE
)Arguments
- replicate_exdf
An
exdfobject representing one CO2 response curve.- fit_options
A list of named elements representing fit options to use for each parameter. Values supplied here override the default values (see details below). Each element must be
'fit','column', or a numeric value. A value of'fit'means that the parameter will be fit; a value of'column'means that the value of the parameter will be taken from a column inreplicate_exdfof the same name; and a numeric value means that the parameter will be set to that value. For example,fit_options = list(RL_at_25 = 0, Vcmax_at_25 = 'fit', Vpmax_at_25 = 'column')means thatRL_at_25will be set to 0,Vcmax_at_25will be fit, andVpmax_at_25will be set to the values in theVpmax_at_25column ofreplicate_exdf.- sd_A
The standard deviation of the measured values of the net CO2 assimilation rate, expressed in units of
micromol m^(-2) s^(-1). Ifsd_Ais not a number, then there must be a column inexdf_objcalledsd_Awith appropriate units. A numeric value supplied here will overwrite the values in thesd_Acolumn ofexdf_objif it exists.- x_etr
The fraction of whole-chain electron transport occurring in the mesophyll (dimensionless). See Equation 29 from S. von Caemmerer (2021).
- a_column_name
The name of the column in
replicate_exdfthat contains the net assimilation inmicromol m^(-2) s^(-1).- ao_column_name
The name of the column in
replicate_exdfthat contains the dimensionless ratio of solubility and diffusivity of O2 to CO2.- ci_column_name
The name of the column in
replicate_exdfthat contains the intercellular CO2 concentration inmicromol mol^(-1).- gamma_star_column_name
The name of the column in
replicate_exdfthat contains the dimensionlessgamma_starvalues.- gmc_norm_column_name
The name of the column in
replicate_exdfthat contains the normalized mesophyll conductance values (with units ofnormalized to gmc at 25 degrees C).- j_norm_column_name
The name of the column in
exdf_objthat contains the normalizedJvalues (with units ofnormalized to J at 25 degrees C).- kc_column_name
The name of the column in
replicate_exdfthat contains the Michaelis-Menten constant for rubisco carboxylation inmicrobar.- ko_column_name
The name of the column in
replicate_exdfthat contains the Michaelis-Menten constant for rubisco oxygenation inmbar.- kp_column_name
The name of the column in
replicate_exdfthat contains the Michaelis-Menten constant for PEP carboxylase carboxylation inmicrobar.- oxygen_column_name
The name of the column in
exdf_objthat contains the concentration of O2 in the ambient air, expressed as a percentage (commonly 21% or 2%); the units must bepercent.- rl_norm_column_name
The name of the column in
replicate_exdfthat contains the normalizedRLvalues (with units ofnormalized to RL at 25 degrees C).- total_pressure_column_name
The name of the column in
exdf_objthat contains the total pressure inbar.- vcmax_norm_column_name
The name of the column in
replicate_exdfthat contains the normalizedVcmaxvalues (with units ofnormalized to Vcmax at 25 degrees C).- vpmax_norm_column_name
The name of the column in
replicate_exdfthat contains the normalizedVpmaxvalues (with units ofnormalized to Vpmax at 25 degrees C).- hard_constraints
To be passed to
calculate_c4_assimilation; see that function for more details.- debug_mode
A logical (
TRUEorFALSE) variable indicating whether to operate in debug mode. In debug mode, information about theguessis printed each time the error function is called; this can be helpful when troubleshooting issues with an optimizer.
Details
When fitting A-Ci curves, it is necessary to define a function that calculates
the likelihood of a given set of alpha_psii, gbs,
gmc_at_25, J_at_25, RL_at_25, Rm_frac,
Vcmax_at_25, Vpmax_at_25, and Vpr values by comparing a
model prediction to a measured curve. This function will be passed to an
optimization algorithm which will determine the values that produce the
smallest error.
The error_function_c4_aci returns such a function, which is based on a
particular A-Ci curve and a set of fitting options. It is possible to just fit
a subset of the available fitting parameters; by default, the fitting
parameters are RL_at_25, Vcmax_at_25, and Vpmax_at_25.
This behavior can be changed via the fit_options argument.
For practical reasons, the function actually returns values of -ln(L),
where L is the likelihood. The logarithm of L is simpler to
calculate than L itself, and the minus sign converts the problem from
a maximization to a minimization, which is important because most optimizers
are designed to minimize a value.
A penalty is added to the error value for any parameter combination where
An is not a number, or where calculate_c4_assimilation
produces an error.
Value
A function with one input argument guess, which should be a numeric
vector representing values of the parameters to be fitted (which are specified
by the fit_options input argument.) Each element of guess is the
value of one parameter (arranged in alphabetical order.) For example, with the
default settings, guess should contain values of RL_at_25,
Vcmax_at_25, and Vpmax_at_25 (in that order).
Examples
# Read an example Licor file included in the PhotoGEA package
licor_file <- read_gasex_file(
PhotoGEA_example_file_path('c4_aci_1.xlsx')
)
# Define a new column that uniquely identifies each curve
licor_file[, 'species_plot'] <-
paste(licor_file[, 'species'], '-', licor_file[, 'plot'] )
# Organize the data
licor_file <- organize_response_curve_data(
licor_file,
'species_plot',
c(9, 10, 16),
'CO2_r_sp'
)
# Calculate temperature-dependent values of C4 photosynthetic parameters
licor_file <- calculate_temperature_response(licor_file, c4_temperature_param_vc)
# Calculate the total pressure in the Licor chamber
licor_file <- calculate_total_pressure(licor_file)
# Define an error function for one curve from the set
error_fcn <- error_function_c4_aci(
licor_file[licor_file[, 'species_plot'] == 'maize - 5', , TRUE]
)
# Evaluate the error for RL_at_25 = 0, Vcmax_at_25 = 35, Vpmax_at_25 = 180
error_fcn(c(0, 35, 180))
#> [1] 140.3377
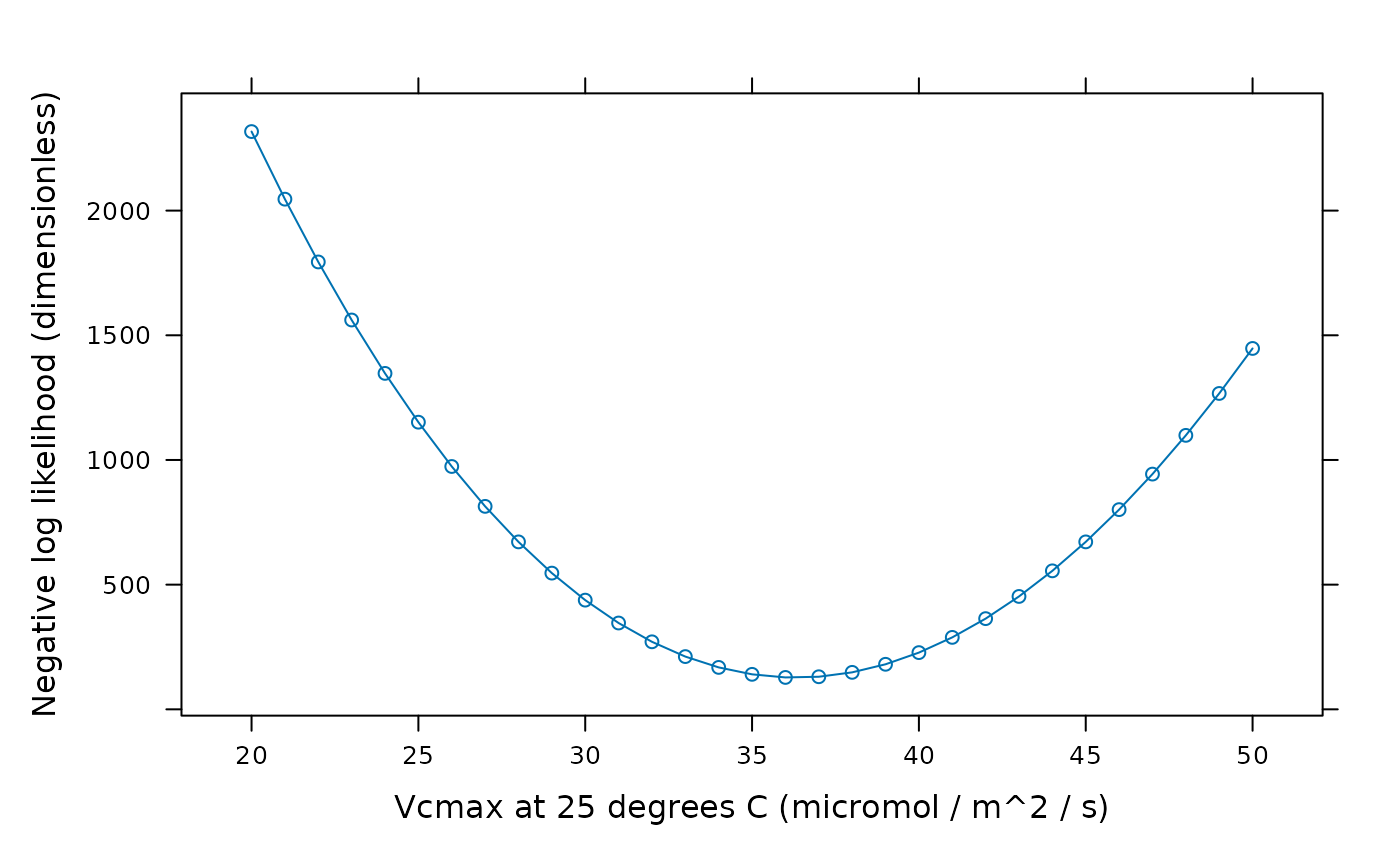
# Make a plot of error vs. Vcmax_at_25 when the other parameters are fixed to
# the values above.
vcmax_error_fcn <- function(Vcmax_at_25) {error_fcn(c(0, Vcmax_at_25, 180))}
vcmax_seq <- seq(20, 50)
lattice::xyplot(
sapply(vcmax_seq, vcmax_error_fcn) ~ vcmax_seq,
type = 'b',
xlab = 'Vcmax at 25 degrees C (micromol / m^2 / s)',
ylab = 'Negative log likelihood (dimensionless)'
)